Multiple I/O
Tasks don't only have to have a single input and output: they can produce accept multiple inputs, produce multiple outputs, and workflows can easily specify which inputs and outputs need to be connected together. For example, consider the following example.
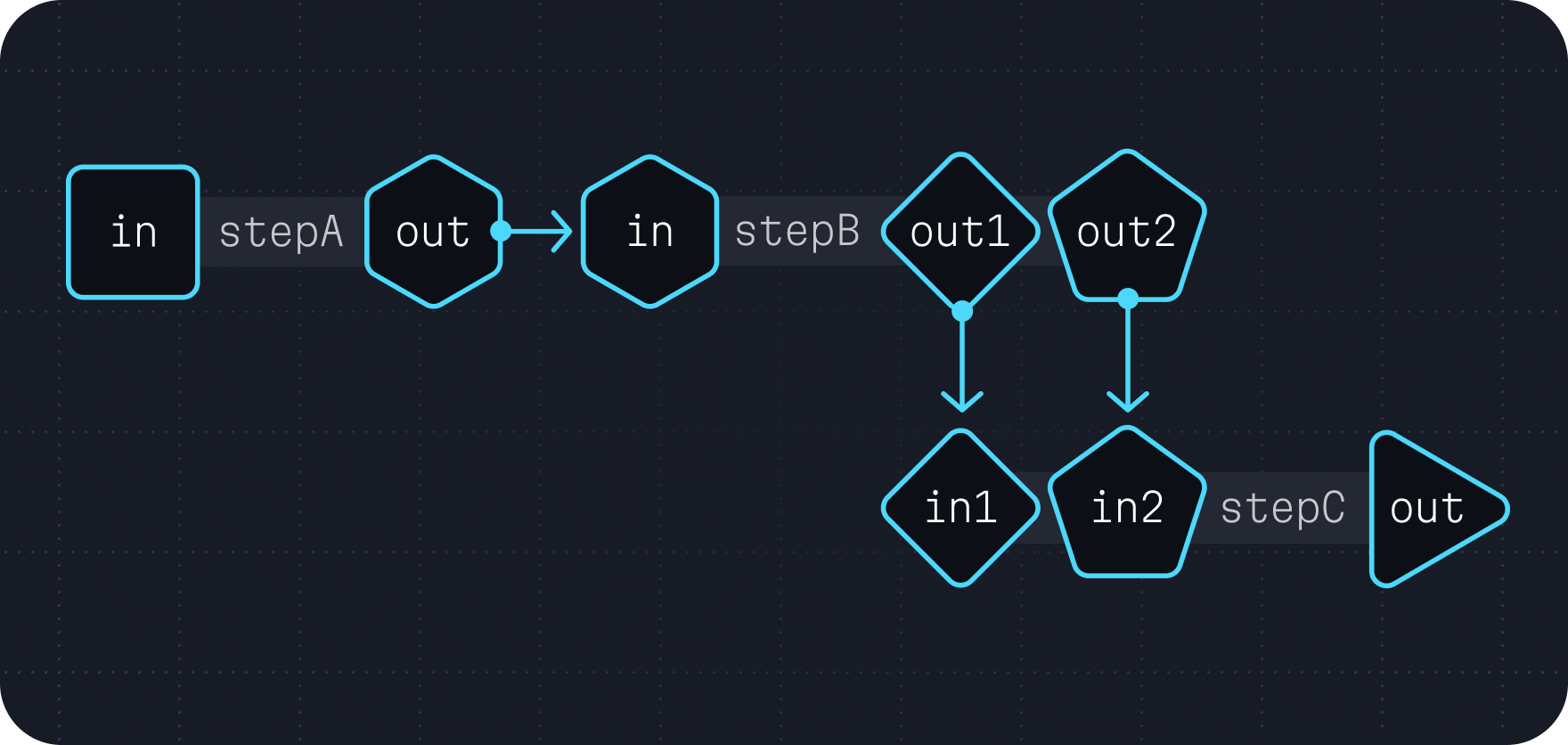
Notice that the two outputs from stepB have distinctive types and names. Further, the two inputs in stepC has distinctive types and names as well. Because of this, you can easily direct a workflow to connect the outputs in stepB to their respective inputs in stepC.
# ... task definitions ...
workflow run {
# Run `stepA`.
call stepA {}
# Run `stepB`, connecting the `in` input to `stepA`'s `out` output.
call stepB { input: in = stepA.out }
# Run `stepC`, connecting the `in1` and `in2` inputs to `stepB`'s
# `out1` and `out2` outputs, respectively.
call stepC { input:
in1 = stepB.out1,
in2 = stepB.out2
}
}Again, because each task creates a scope containing each output, the correct inputs and outputs can be connected together, and names will not conflict.
An example
To illustrate this concept, here is an example workflow that uses Picard to split a VCF into (a) a VCF containing only single nucleotide variants and (b) a VCF containing only insertions/deletions, followed by merging them back together.
version 1.3
# (1) We write a task to split the VCF into multiple VCFs.
task split_vcf {
input {
# (a) The task takes in a single input VCF.
File vcf
}
command <<<
java -jar picard.jar SplitVcfs \
I="~{vcf}" \
SNP_OUTPUT="snp.vcf" \
INDEL_OUTPUT="indel.vcf" \
STRICT=false
>>>
output {
# (b) The task produces two output VCFs.
File snp_vcf = "snp.vcf"
File indel_vcf = "indel.vcf"
}
}
# (2) We write a task to merge these two VCFs.
task merge_vcfs {
input {
# (a) Two input VCFs are accepted.
File snp_vcf
File indel_vcf
}
command <<<
java -jar picard.jar MergeVcfs \
I="~{snp_vcf}" \
I="~{indel_vcf}" \
O="combined.vcf"
>>>
output {
# (b) One output VCF is produced.
File combined = "combined.vcf"
}
}
# (3) We write a workflow that connects the multiple outputs
# from the `split_vcf` task into the multiple inputs from the
# `merge_vcfs` task.
workflow run {
input {
File vcf
}
call split_vcf { input: vcf }
# (a) The outputs are connected to the inputs here.
call merge_vcfs { input:
snp_vcf = split_vcf.snp_vcf,
indel_vcf = split_vcf.indel_vcf,
}
output {
File combined = merge_vcfs.combined
}
}